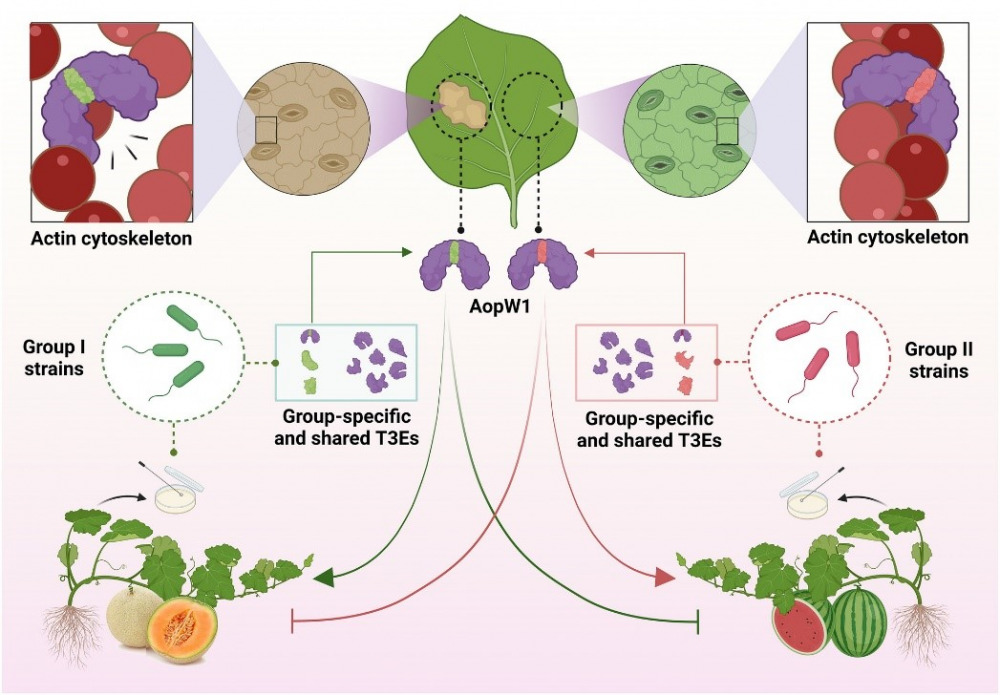
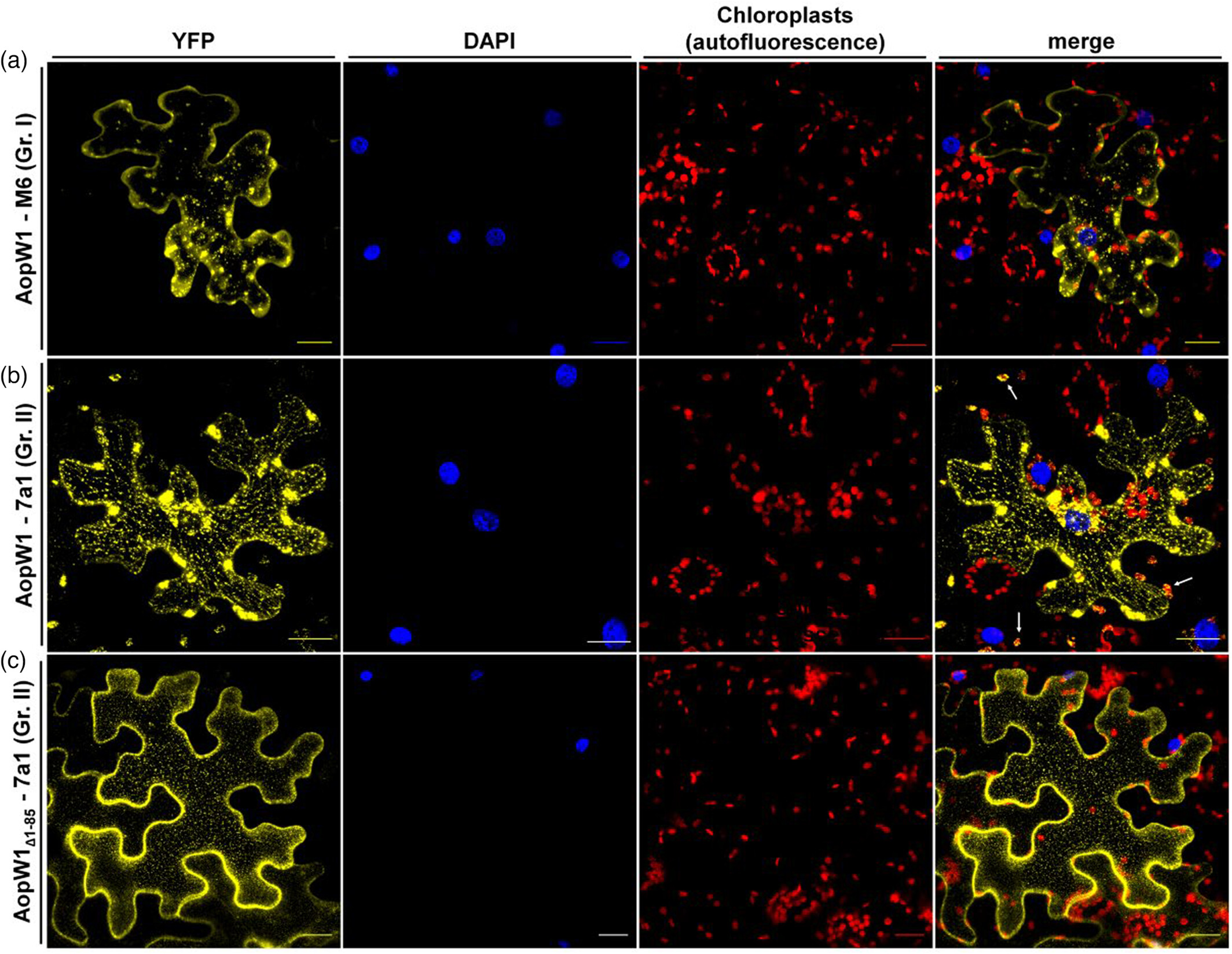
Our new findings from the Burdman lab were featured in the Hebrew University and Jerusalem Post. Researchers: Irene Jiménez-Guerrero, Monica Sonawane, Noam Eckshtain-Levi, Za Khai Tuang, Gustavo Mateus da Silva, Francisco Pérez-Montaño, Meirav Leibman-Markus, Rupali Gupta, Lianet Noda-Garcia, Maya Bar, Saul Burdman; In this work, we focused on the effector AopW1, revealing its role in host adaptation and providing new insights into the HopW1 family of bacterial effectors, with the abstract of this paper accessible here. Acidovorax citrulli, the bacterium that causing disease to melon and watermelon, has two major genetic groups with different host preferences. Group I strains are more associated with melons, while Group II strains are more aggressive towards watermelons. We discovered that these groups differ in their arsenal of type-III secreted protein effectors, which are crucial in manipulating host metabolism to promote disease. Our study sheds light on how these effectors, particularly AopW1, contribute to the pathogen’s virulence and host preference. This knowledge is vital for developing strategies to combat bacterial fruit blotch and protect melon and watermelon crops.
Natural variation in a short region of the Acidovorax citrulli type III-secreted effector AopW1 is associated with differences in cytotoxicity and host adaptation
Natural variation in a short region of the Acidovorax citrulli type III-secreted effector AopW1 is associated with differences in cytotoxicity and host adaptation. Abstract: Bacterial fruit blotch, caused by Acidovorax citrulli, is a serious disease of melon and watermelon. The strains of the pathogen belong to two major genetic groups: group I strains are strongly associated with melon, while group II strains are more aggressive on watermelon. A. citrulli secretes many protein effectors to the host cell via the type III secretion system. Here we characterized AopW1, an effector that shares similarity to the actin cytoskeleton-disrupting effector HopW1 of Pseudomonas syringae and with effectors from other plant-pathogenic bacterial species. AopW1 has a highly variable region (HVR) within amino acid positions 147 to 192, showing 14 amino acid differences between group I and II variants. We show that group I AopW1 is more toxic to yeast and Nicotiana benthamiana cells than group II AopW1, having stronger actin filament disruption activity, and increased ability to induce cell death and reduce callose deposition. We further demonstrated the importance of some amino acid positions within the HVR for AopW1 cytotoxicity. Cellular analyses revealed that AopW1 also localizes to the endoplasmic reticulum, chloroplasts, and plant endosomes. We also show that overexpression of the endosome-associated protein EHD1 attenuates AopW1-induced cell death and increases defense responses. Finally, we show that sequence variation in AopW1 plays a significant role in the adaptation of group I and II strains to their preferred hosts, melon and watermelon, respectively. This study provides new insights into the HopW1 family of bacterial effectors and provides first evidence on the involvement of EHD1 in response to biotic stress. Read the full article here: https://doi.org/10.1111/tpj.16507
Al Nakba
Al Nakba cih kammal pen Zomite sungah a thei ngei leh a za ngei kitam lo kha ding hi. Hih kammal a kizat cilna pen Israel leh Palestiniante kibuaina nasia takinin omin, Palestiniante mi 750,000 bang asi amang a om hun, 1948 kum ahi hi. Tu laitak aa Israel leh Hamas (Palestinian galkapte-gongtatte) a kidona uh ahang tampi om ding sung panin, a tawsawn a thupi khat pen, gam (leitang) kituh uh ahi hi. Ahih leh, tua a kituh uh leitang (gam) kua aa’ taktak hizaw ding? Tanglai a’ thupiang Al Nakba hong phawk kik sak hi. Hih article a kicingin Zomi History official website ah kisim thei hi. Tua Al Nakba tawh kizomin apiang toto tulai tak aa, Israel leh Palestine buainate tawh kisai-in Gog le Magog peuh ahi kha tam cih ngaihsutna tawh kisai, himun panin kisim thei ding hi.
Israel – Hamas Kibuaina gam vaiah Israel – Palestine kuate Innteek zaw uh hiam!
Author: Tuang Za Khai; Israel – Hamas Kibuaina gam vaiah Israel – Palestine kuate Innteek zaw uh hiam! cih thu ih ngaihsut ciangin, laibu khempeuh lak panin Lai Siangtho bek mah a kician penin etkak theih dingin a om suak hi. Israel tawh kisai-in, Piancil 11:28, 11:31 leh 15:7 te sim lehang Israelte’ pianna pupi Abraham pen Chaldees gam Ur khua-ah piangin khang khia-ahih lam kilangh hi. Pasianin Abraham hopih in tua gam nusia henla hileh, a suan akhak thupha tampi pia ding ahihna gen hi (Pian. 12:1-2). Abrahamin Pasian sapna mangin tua gam nusia in Canaan gam tung hi (Pian. 12:5). Tua laitak Canaan gam ah Hittites, Amorites, leh Canaanites mite teng uh hi (Pian. 12:6; 15:18-21). Tua mun mah ahi Hebron a kicina mun ah Abraham in a zi Sarah na phum aa (Pian. 23:19), amah zong kum 175 bang aphak nungah si-in, a zi tawh kiphum khawm hi (Pian. 25:7-10). Hithu a kicingin Tonsan News ah kisim thei ding hi. Thamlo in hih buaina tung tawnin hun nunung tawh a kizomzomta ahitam, cih tawh kisai-in muhna tuamtuam siksanin a kigelh thu himun panin kisim thei hi.
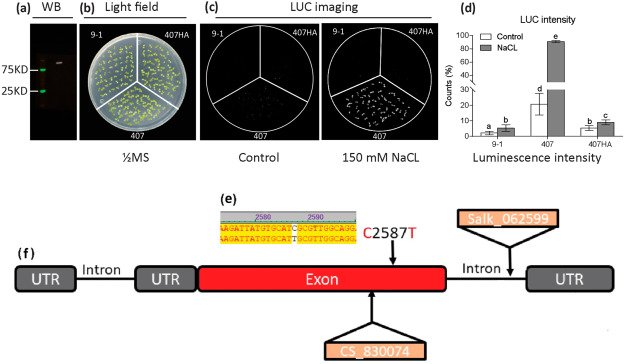
Preliminary analysis reveals that RCF1 confers resistance to Pseudomonas syringae pv. tomato DC3000 but impairs Botrytis cinerea infection
Author: Za Khai Tuang, Tial C. Ling; The adaptation process is a way of life for plants, and they must contend with biotic and abiotic stresses for their chance of survival. These stresses can lead to disastrous crop production and limited distribution of crops geographically. The use of chemicals to address those stresses in agriculture is limited, while management strategies often fail to meet the needs of the industry. Therefore, novel techniques are needed to control plant diseases and to maintain food security. RCF1 has been found to encode an RNA helicase containing DEAD-box domains and loss-of-function mutation of this gene increases the sensitivity of plants to cold stress. In this study, a single mutant generated in RCF1 was used to investigate the function of the RCF1 gene in pathogen infection stress (i.e., Pst DC3000 and B. cinerea). RFC1 was found to confer resistance to Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) but negatively affect Botrytis cinerea (B. cinerea) infection by repressing SNI1 pathways and JA signaling while activating SA pathways and its receptor NPR1. The generated rcf1 allelic mutant (name: 407) showed sensitivities to bacteria pathogen (Pst DC3000) but tolerance to fungus infection (B. cinerea), and SNI1 was highly upregulated in it. Free and conjugated Salicylic Acid (SA) contents were much lower in the allelic mutant 407 than in complementary materials and is sensitive to Pst DC3000 infection, and NPR1 depending on SA biosynthetic genes were down-regulated in it. Our preliminary results reveal that RCF1 confers resistance to Pst DC3000 by SA and NPR1 dependence manner but blocks SNI1 and JA pathways. Read the full article by clicking here.
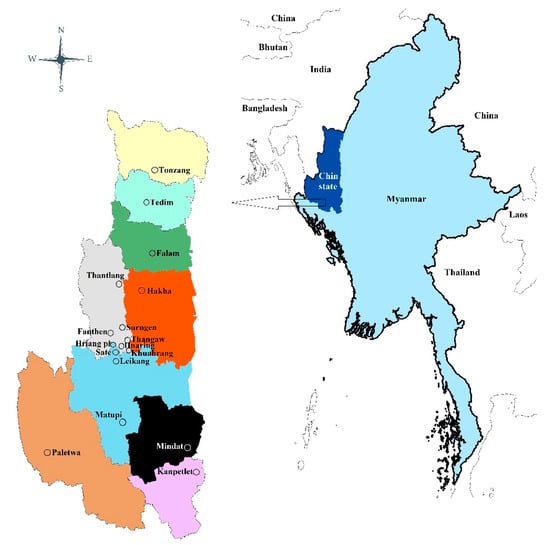
Traditional Knowledge of Textile Dyeing Plants: A Case Study in the Chin Ethnic Group of Western Myanmar
Authors: Tial C. Ling, Angkhana Inta, Kate E. Armstrong, Damon P. Little,Pimonrat Tiansawat,Yong-Ping Yang, Patcharin Phokasem, Za Khai Tuang, Chainarong Sinpoo, Terd Disayathanoowat; Traditional knowledge of the plants used for textile dyeing is disappearing due to the utilization of synthetic dyes. Recently, natural products made from plants have gained global interest. Thus, preserving traditional knowledge of textile dyeing plants is crucial. Here, we documented this knowledge by interviewing 2070 informants from 14 communities of the Chin ethnic group of Myanmar. The Chin communities we interviewed used a total of 32 plant species for textile dyeing from 29 genera in 24 families. Chromolaena odorata, Lithocarpus fenestratus, and L. pachyphyllus were the most important dye species. The most common responses described dyes that were red in color, produced from leaves, derived from tree species, collected from the wild, and used as firewood ash as a mordant to fix the dye to the fabrics. According to the IUCN Red List of threatened species, one species was registered as Data Deficient, 20 species still needed to be categorized, and 11 species were categorized as Least Concern. This study will help re-establish the use of natural dyes, encourage the cultural integrity of the indigenous people, and serve as an example for other communities to preserve their traditional knowledge of plant textile dyes. Textile: Material made from fibers and yarns, originally referring to woven fabrics. Now encompasses a wide range of products including knitted, tufted, and non-woven materials used in clothing, home furnishings, and industrial applications. Read the full article here.